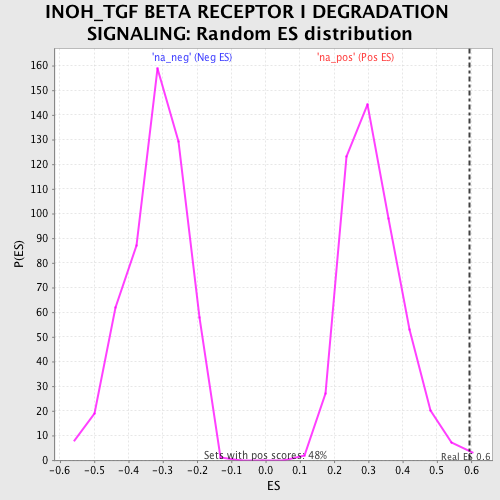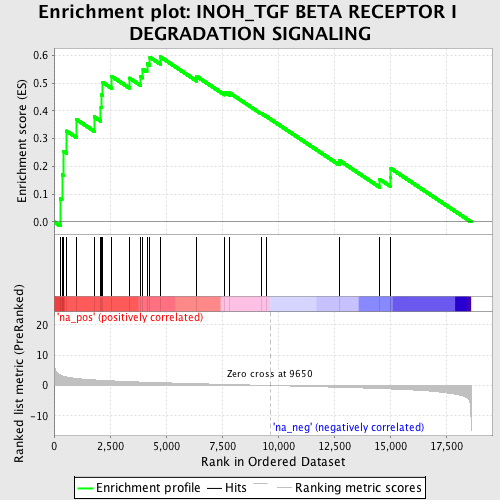
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | INOH_TGF BETA RECEPTOR I DEGRADATION SIGNALING |
| Enrichment Score (ES) | 0.59468895 |
| Normalized Enrichment Score (NES) | 1.901921 |
| Nominal p-value | 0.004192872 |
| FDR q-value | 0.031849366 |
| FWER p-Value | 0.393 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | PSMB1 | 271 | 3.438 | 0.0838 | Yes | ||
| 2 | TGFBR1 | 354 | 3.193 | 0.1707 | Yes | ||
| 3 | PSMB6 | 406 | 3.058 | 0.2555 | Yes | ||
| 4 | PSMB10 | 554 | 2.796 | 0.3276 | Yes | ||
| 5 | PSMB9 | 1005 | 2.291 | 0.3690 | Yes | ||
| 6 | PSMB2 | 1796 | 1.824 | 0.3787 | Yes | ||
| 7 | PSMB3 | 2090 | 1.688 | 0.4112 | Yes | ||
| 8 | PSMA5 | 2104 | 1.682 | 0.4586 | Yes | ||
| 9 | PSMB8 | 2176 | 1.657 | 0.5022 | Yes | ||
| 10 | PSMA3 | 2575 | 1.524 | 0.5245 | Yes | ||
| 11 | BMPR1A | 3362 | 1.271 | 0.5186 | Yes | ||
| 12 | PSMA4 | 3845 | 1.130 | 0.5249 | Yes | ||
| 13 | PSMB5 | 3966 | 1.100 | 0.5500 | Yes | ||
| 14 | ACVRL1 | 4147 | 1.059 | 0.5706 | Yes | ||
| 15 | PSMA6 | 4276 | 1.032 | 0.5932 | Yes | ||
| 16 | PSMD2 | 4743 | 0.926 | 0.5947 | Yes | ||
| 17 | PSMA2 | 6367 | 0.596 | 0.5245 | No | ||
| 18 | SMAD7 | 7618 | 0.363 | 0.4676 | No | ||
| 19 | PSMB4 | 7842 | 0.322 | 0.4648 | No | ||
| 20 | PSMA7 | 9241 | 0.077 | 0.3919 | No | ||
| 21 | PSMB7 | 9486 | 0.035 | 0.3797 | No | ||
| 22 | BMPR1B | 12733 | -0.587 | 0.2219 | No | ||
| 23 | PSMA1 | 14534 | -1.013 | 0.1541 | No | ||
| 24 | PSMC1 | 15017 | -1.140 | 0.1608 | No | ||
| 25 | ACVR1 | 15033 | -1.143 | 0.1927 | No |